
3D Metabolic Networks
|
|
|
|
|
|
|
|
|
|
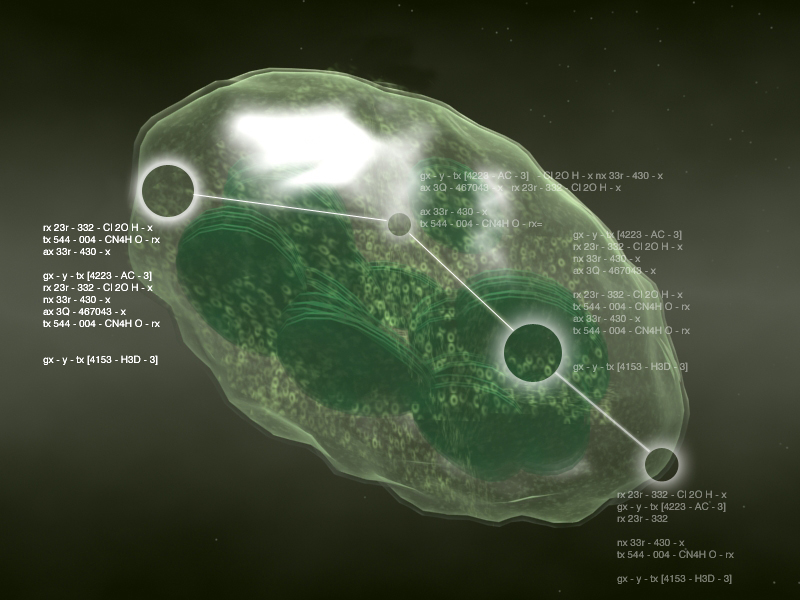 |
 |
| 3D Diagram of a chloroplast with embedded metabolic information (Artist: Andres Reinot) | Metabolic network with navigation controlled by a tablet computer controlled by Kris Blom. |
Description
Multidimensional genome-wide gene expression profiling datasets are being generated for bacterial, plant and animal systems. This complex data reveals the interacting metabolic pathways that control cell metabolism. Ultimately, understanding these pathways will increase our ability to predict the effects of a given drug on human metabolism, the consequences of changes in a single gene on the composition of a seed, or the effect of a given mutation in a pre-cancer cell. Traditional biology research and analysis methods cannot evaluate the information in these large datasets. This project will integrate advanced biological knowledge with automated mathematical logic, complex data structures, fuzzy cognitive maps, interactive graph visualization, and other computational tools to create a novel analytical suite of tools for metabolic pathway analysis and visualization.
We will visualize and model complex metabolic pathways, developing a software system that enables scalable immersive environments, which will range from desktop visualization to the NSF-supported fully immersive C6 projection system in the Virtual Reality Applications Center (VRAC) at Iowa State. This project will lead to new methods for displaying and navigating through complex metabolic networks in three dimensions. The visual methods will be complimented by existing modeling methods that use fuzzy cognitive maps to analyze network interactions and determine their agreement with the experimental datasets.
Two experimental applications will be developed. First, a prototype interactive visualization of a metabolic network, the Calvin cycle of photosynthesis, will be used to create a living dynamic example of a biochemical pathway for K-12 students to explore. The pathway will be brought alive by a computer game that students explore at their own pace, with rewards for particular achievements such as combining the proper molecules. Complex concepts such as the conversion of chemical, light and heat energy, metabolic flux, use and synthesis of chemical constituents of living organisms can all be illustrated through this pathway. Secondly, three-dimensional visualization will be applied to the analysis of the acetyl-CoA metabolic network in the genetic model-plant Arabidopsis. This visualization will be integrated with complex datasets taken from a group of genetic mutants in specific steps in this pathway.
Project Personnel
- Julie A Dickerson (PI) Electrical and Computer Engineering
- Carolina Cruz-Neira (co-PI) Industrial Engineering
- Eve Wurtele (co-PI) Botany
- Yuting Yang (PhD student) Computer Engineering
- Kris Blom (PhD student) Computer Engineering
- Andres Reinot (undergraduate) Computer Engineering
Publications
- Dickerson JA, Yang Y, Blom K, Reinot A, Lie J, Cruz-Neira C, Wurtele ES. 2003. "Using Virtual Reality to Understand Complex Metabolic Networks." Atlantic Symposium on Computational Biology and Genomic Information Systems and Technology, September. In press.
Funding Sources:
National Science Foundation, Information Technology Research Program, Division of Experimental and Integrative Biology. (Grant # 0219366)
Iowa State University Carver Grant for Explorative Research

![]()